
Helper-Functions Guide
helper-functions.RmdThis guide walks through the use of: get_sigma_post(), get_mu_preds(), get_tau_preds(), get_y_preds(), and get_ate_post(). These are helper functions to make accessing important estimates easier without the need to interact directly with the model output.
Create a Fitted Model
#Load the mvbcf package
library(mvbcf)
#set seed
set.seed(101)
#Create some synthetic data
#number of observations
n<-500
#number of covariates
p<-5
#matrix of covariates
X<-matrix(runif(n*p), nrow=n)
#The outcome under control for y1 and y2
mu1<-3*X[,1]-4*X[,3]^2
mu2<-4*X[,1]+1*X[,3]^2-2*X[,5]
#The effect of receiving treatment on y1 and y2
tau1<-1*X[,1]
tau2<-0.6*X[,1]
#The probability of receiving treatment (true propensity score)
true_propensity<-(X[,2]+X[,3])/3
#Treatment status
Z<-rbinom(n, 1, true_propensity)
#The observed outcomes
y1<-mu1+Z*tau1+rnorm(n, 0, 1)
y2<-mu2+Z*tau2+rnorm(n, 0, 1)
Y<-cbind(y1, y2)
mvbcf_mod <- run_mvbcf(X,
Y,
Z,
X)get_sigma_post()
The get_sigma_post() function accesses the posterior of where and are the specified first and second outcome variables. If and are the same outcome, this corresponds to the residual variance of outcome . If and are different outcome variables, this is the residual covariance of outcomes and .
y1_residual_variance<-get_sigma_post(mvbcf_mod,
1,
1)
y1_y2_residual_covariance<-get_sigma_post(mvbcf_mod,
1,
2)
plot(y1_residual_variance, type="l", ylab="Sigma11 Samples")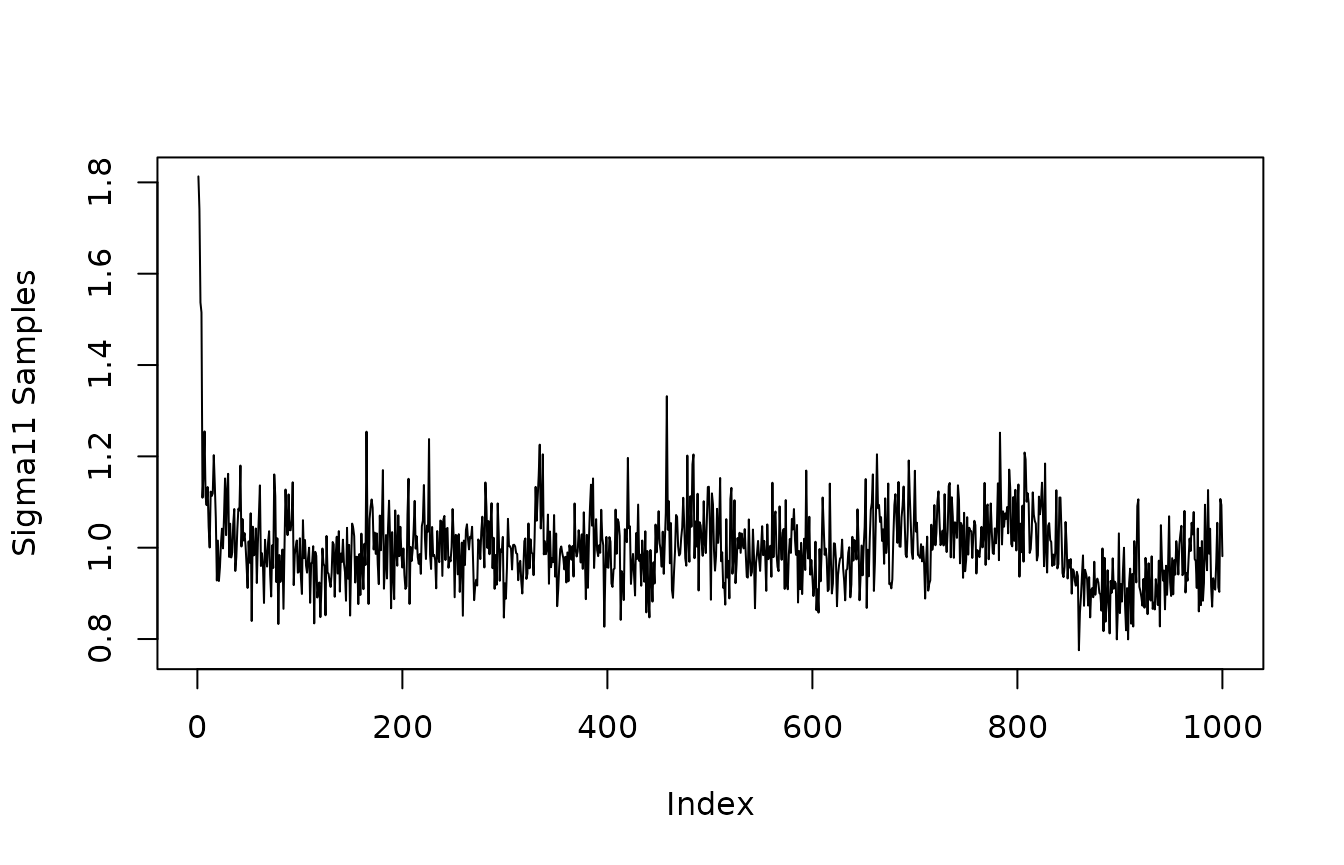
plot(y1_y2_residual_covariance, type="l", ylab="Sigma12 Samples")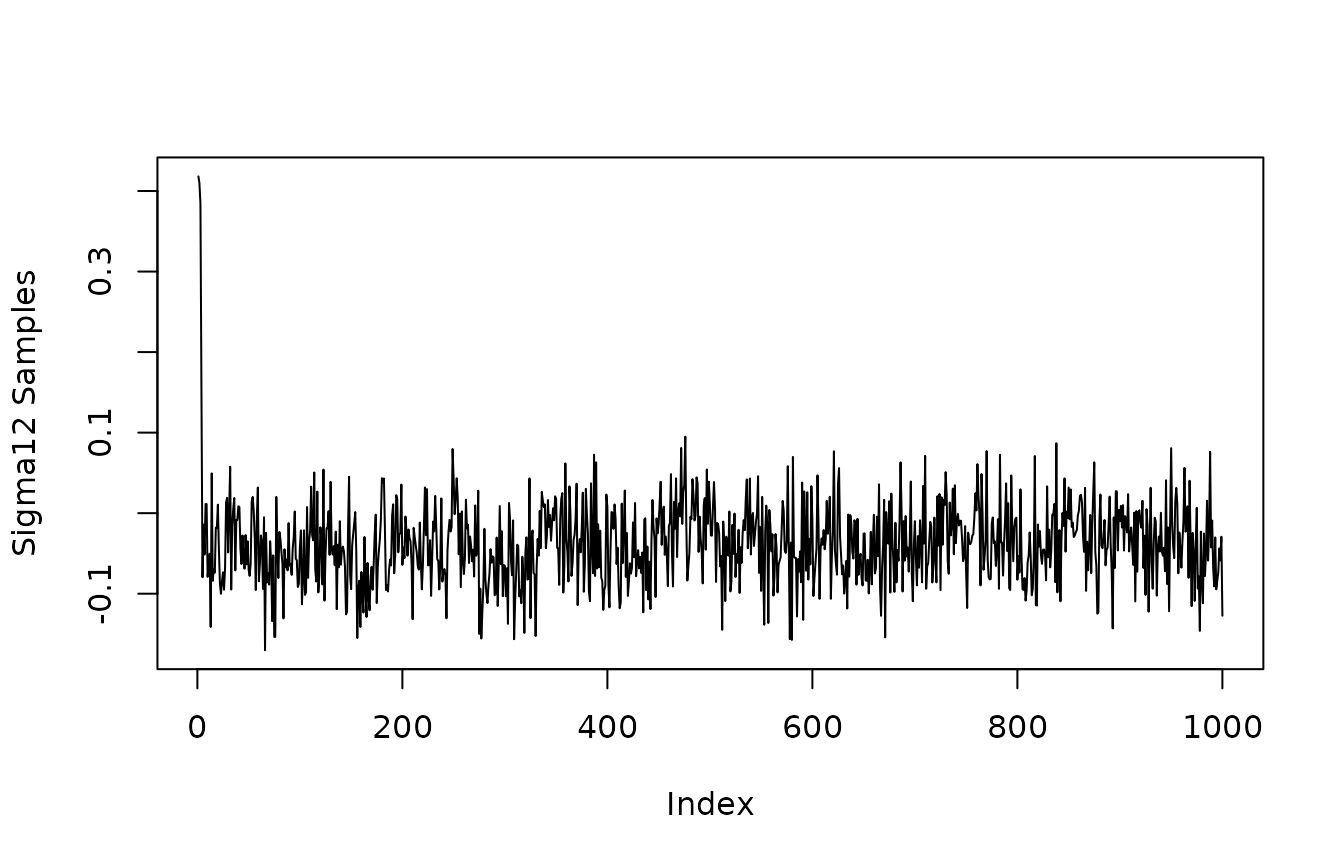
get_tau_preds()
This function is for accessing the individual conditional average treatment effects (ICATEs). You need to specify the outcome variable the ICATEs correspond to, and whether or not the estimates should correspond to the train or test data.
y1_tau_preds<-get_tau_preds(mvbcf_mod,
1,
"train")
y2_tau_preds<-get_tau_preds(mvbcf_mod,
2,
"train")
plot(tau1, y1_tau_preds, main="Predicted vs. True Values", xlab="Tau1", ylab="Tau1 Predictions")
abline(0,1)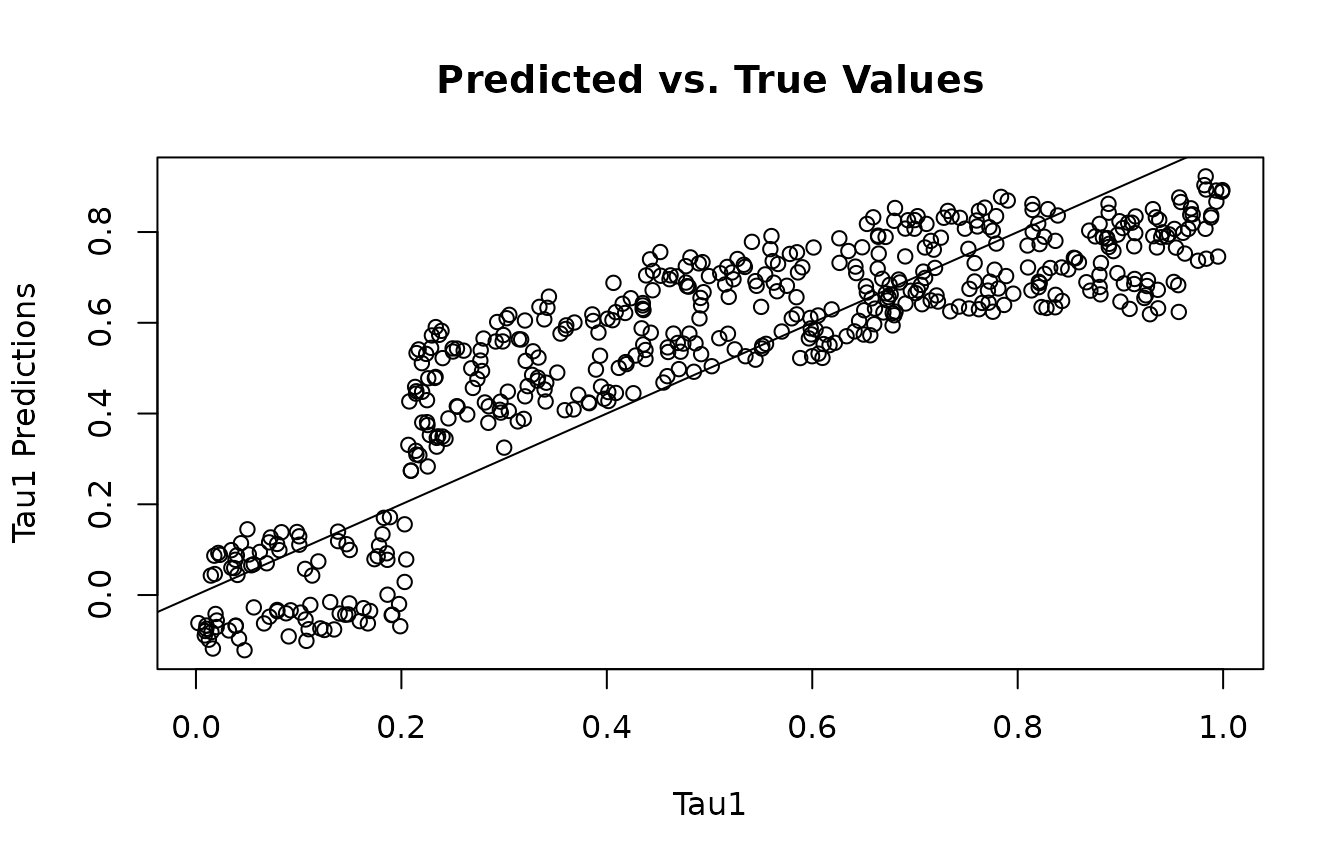
plot(tau2, y2_tau_preds, main="Predicted vs. True Values", xlab="Tau1", ylab="Tau1 Predictions")
abline(0,1)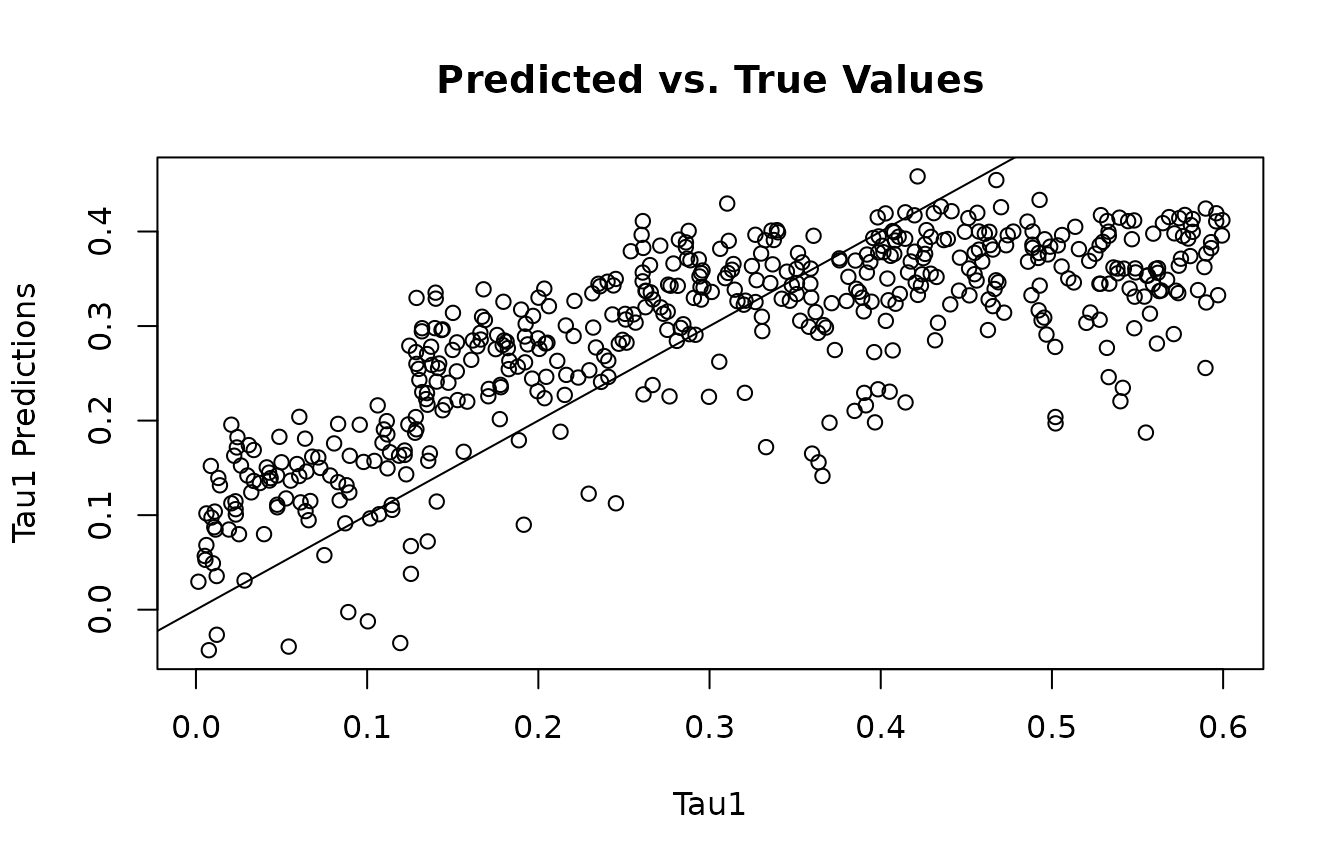
get_mu_preds()
This function is for accessing the individual mu (prognostic effect) predictions. You need to specify the outcome variable the estimates correspond to, and whether or not the estimates should correspond to the train or test data.
y1_mu_preds<-get_mu_preds(mvbcf_mod,
1,
"train")
y2_mu_preds<-get_mu_preds(mvbcf_mod,
2,
"train")
plot(mu1, y1_mu_preds, main="Predicted vs. True Values", xlab="Mu1", ylab="Mu1 Predictions")
abline(0,1)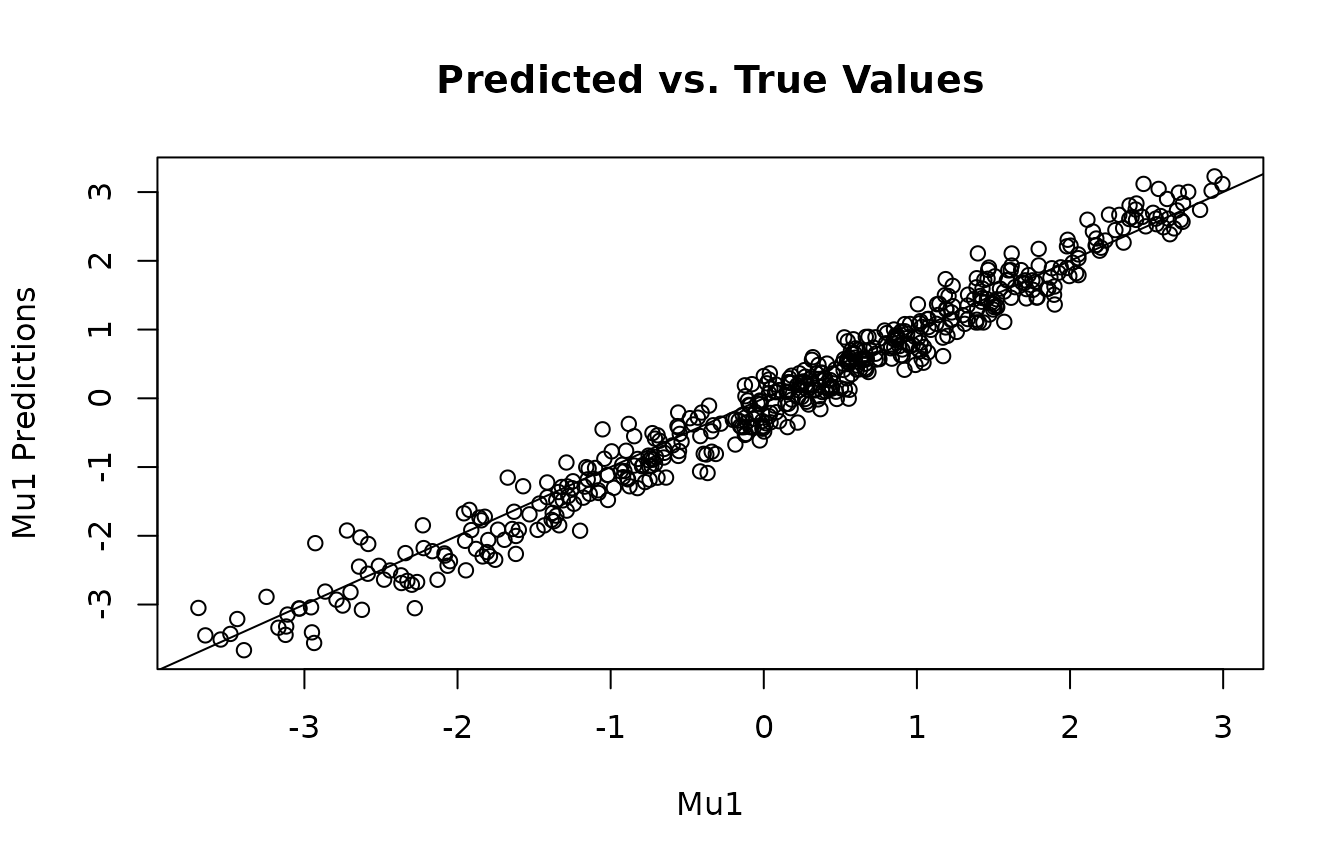
plot(mu2, y2_mu_preds, main="Predicted vs. True Values", xlab="Mu1", ylab="Mu1 Predictions")
abline(0,1)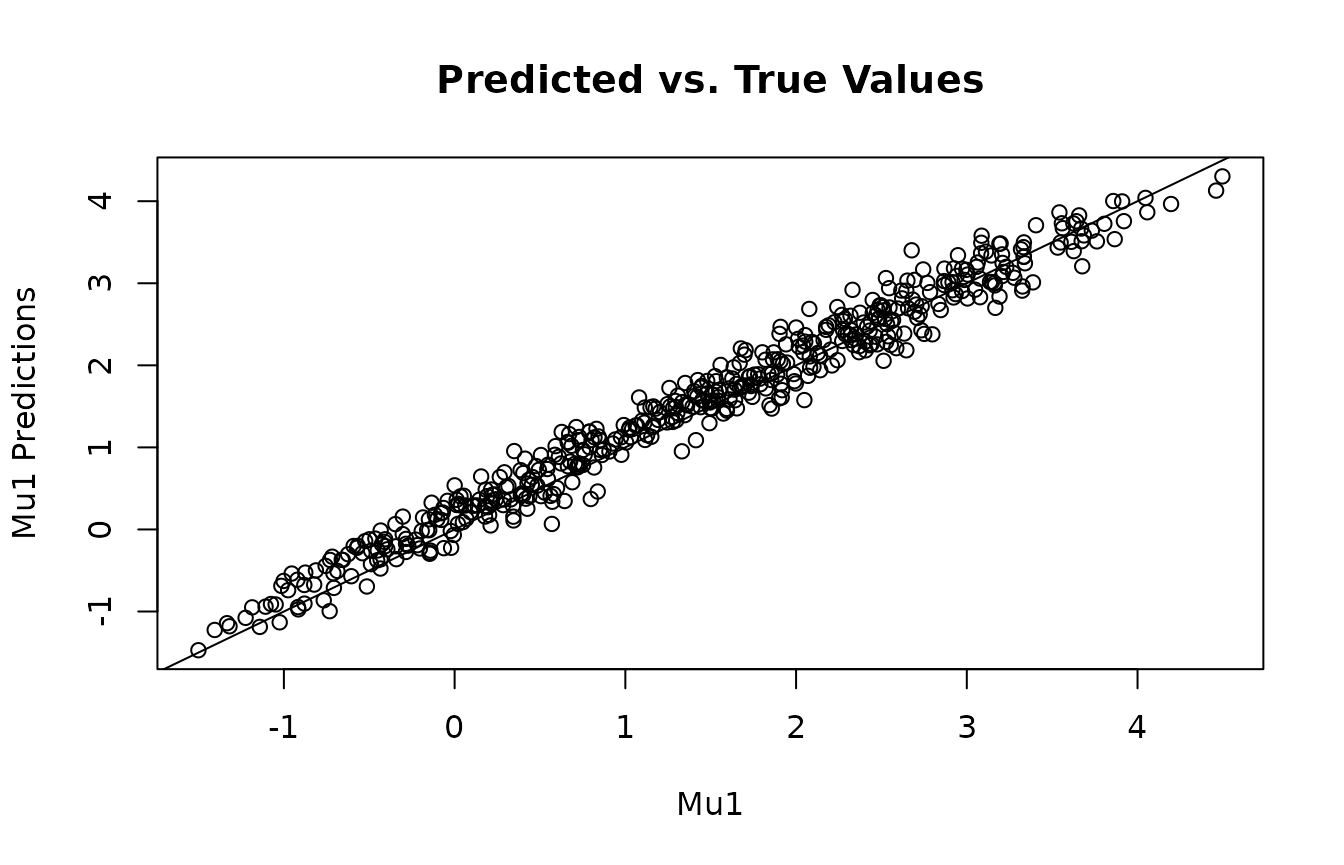
get_y_preds()
This function is for accessing the individual y (mu+Z*tau) predictions. You need to specify the outcome variable the estimates correspond to, and whether or not the estimates should correspond to the train or test data.
y1_preds<-get_y_preds(mvbcf_mod,
1,
"train",
Z)
y2_preds<-get_y_preds(mvbcf_mod,
2,
"train",
Z)
plot(y1, y1_preds, main="Predicted vs. True Values", xlab="Y1", ylab="Y1 Predictions")
abline(0,1)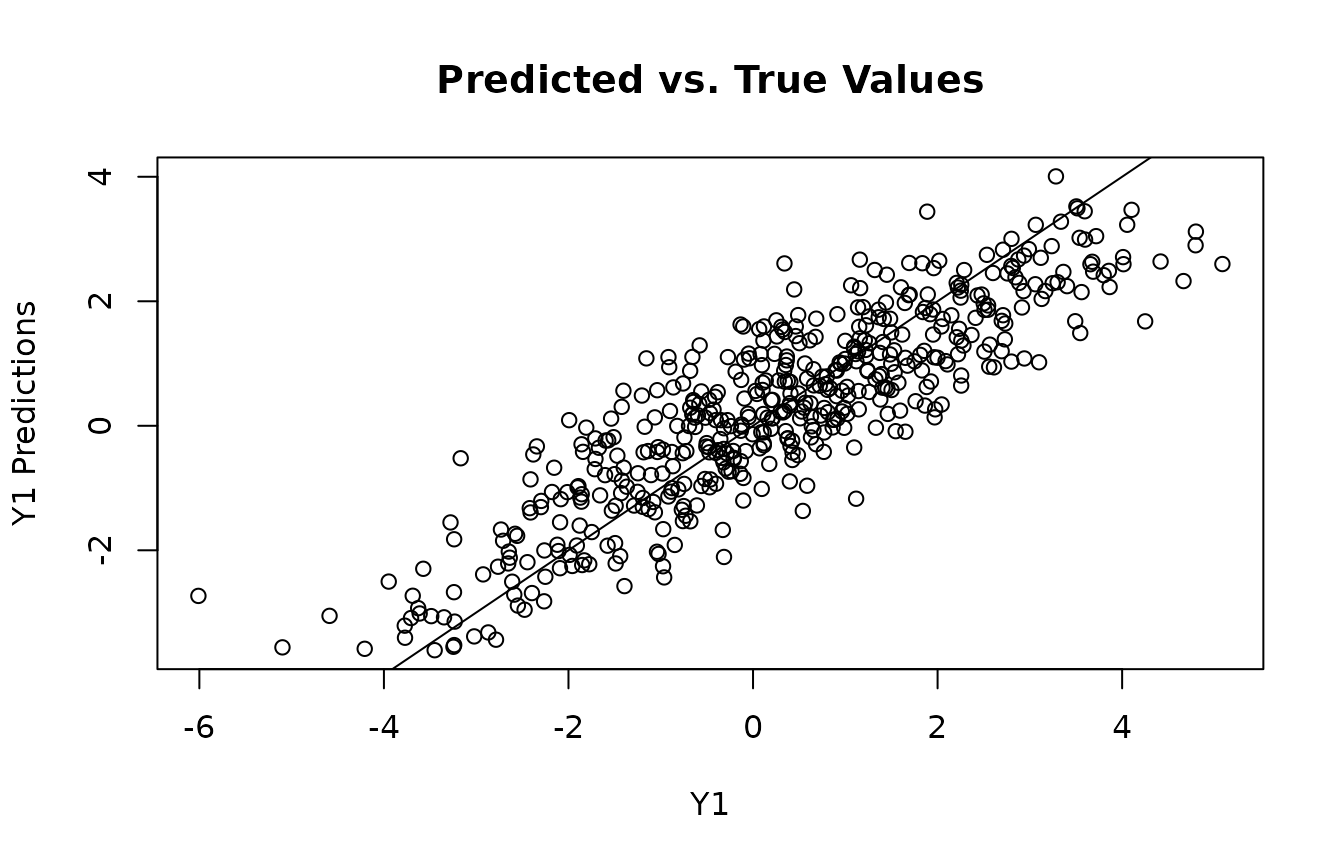
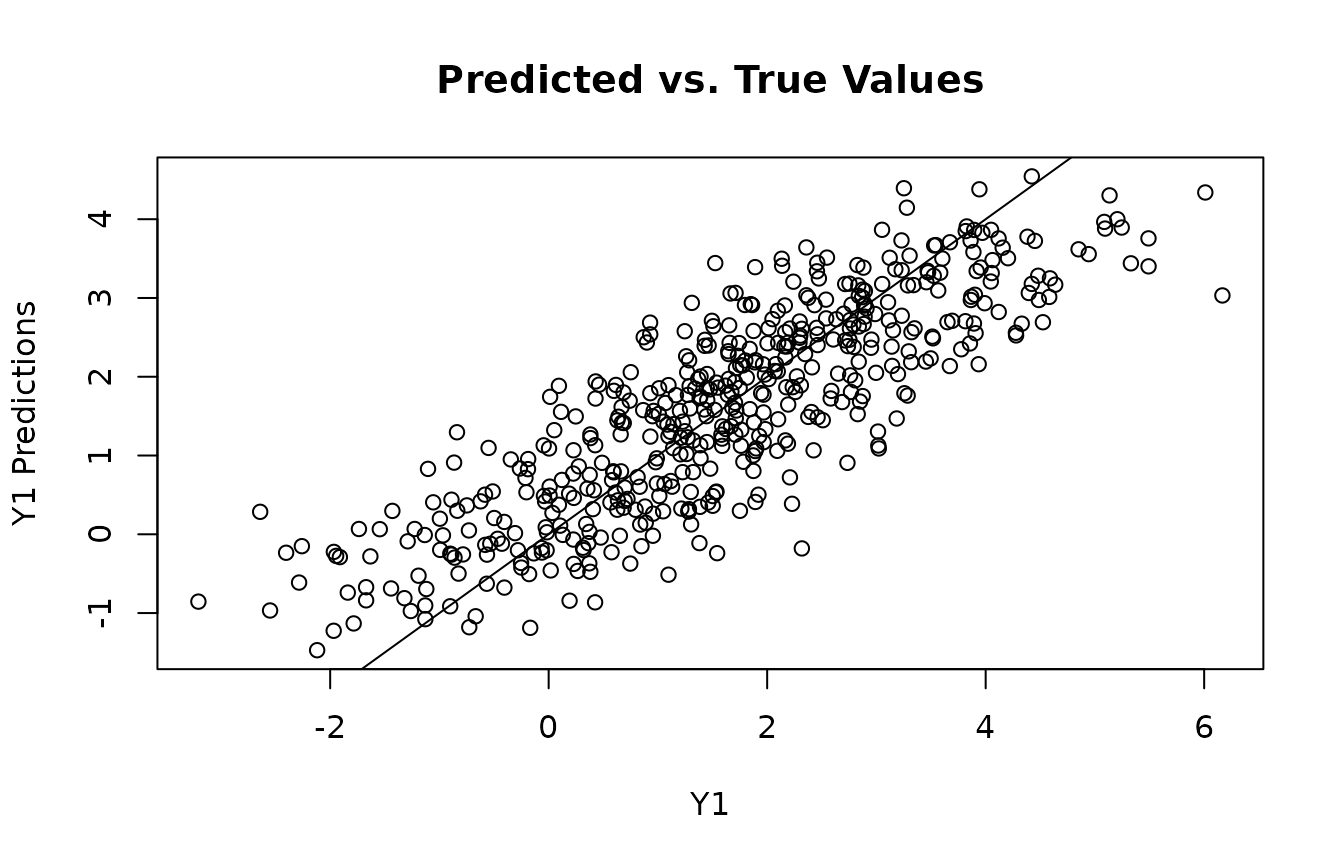
get_ate_post()
This function is for accessing the posterior distribution of the average treatment effect (ATE), produced by averaging the ICATEs for all observations over all iterations of the MCMC sampler. You need to specify the outcome variable the estimates correspond to, and whether or not the estimates should correspond to the train or test data.
y1_ate_post<-get_ate_post(mvbcf_mod,
1,
"train")
y2_ate_post<-get_ate_post(mvbcf_mod,
2,
"train")
plot(density(y1_ate_post), xlab="ATE Outcome 1", main="Posterior of ATE for Outcome 1")
